We’re excited to present our newest paper, currently in preprint: T cell correction pipeline for Inborn Errors of Immunity.
Abstract
CRISPR/Cas9 gene editing technology is a promising tool for correcting pathogenic variants for autologous cell therapies for Inborn Errors of Immunity (IEI). The present IEI correction strategies mainly focus on the knock-in of therapeutic cDNAs, or knockout of the disease-causing gene when feasible. These strategies address many single-gene defects but may disrupt gene expression and require significant optimization for each newly discovered IEI-causing gene, highlighting the need for complementary platforms that can precisely correct diverse pathogenic variants.
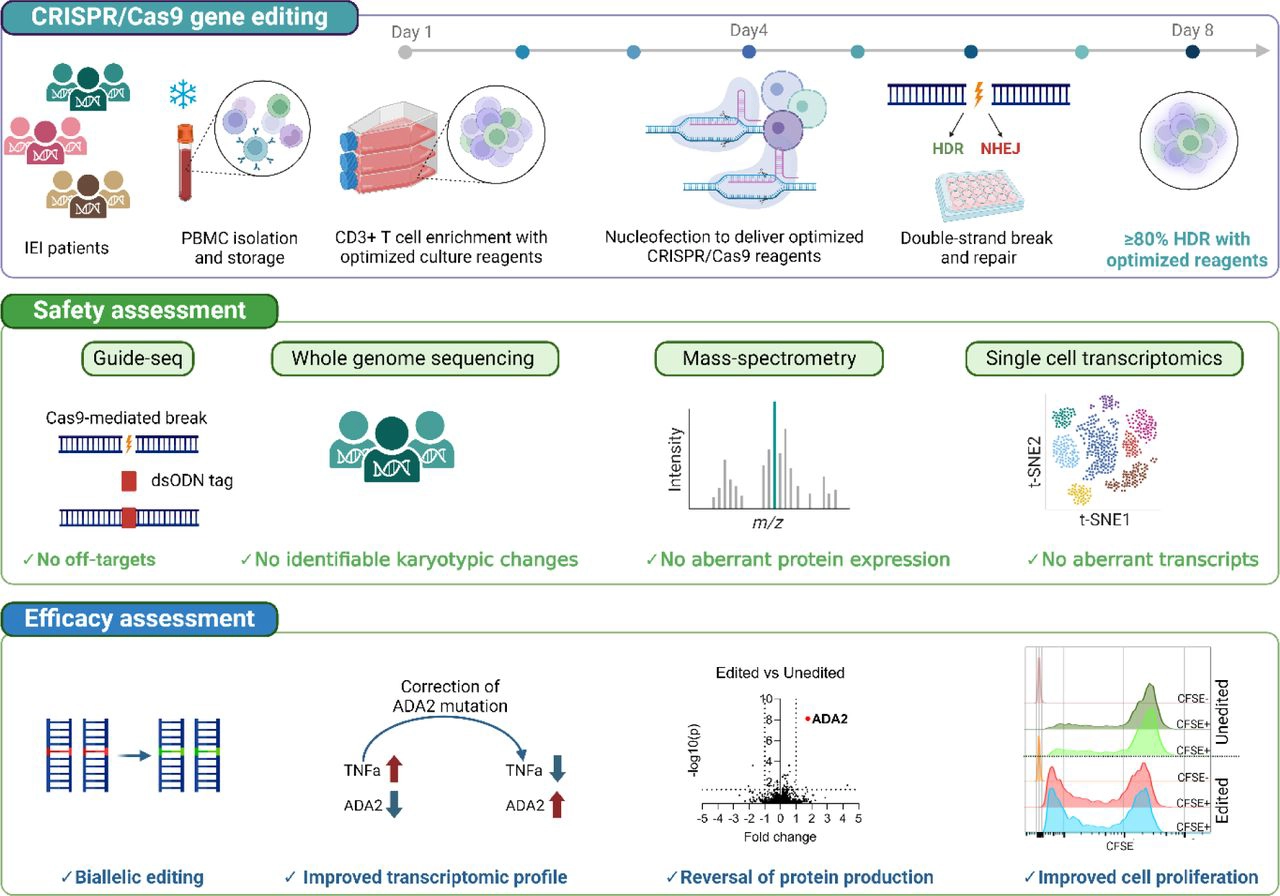
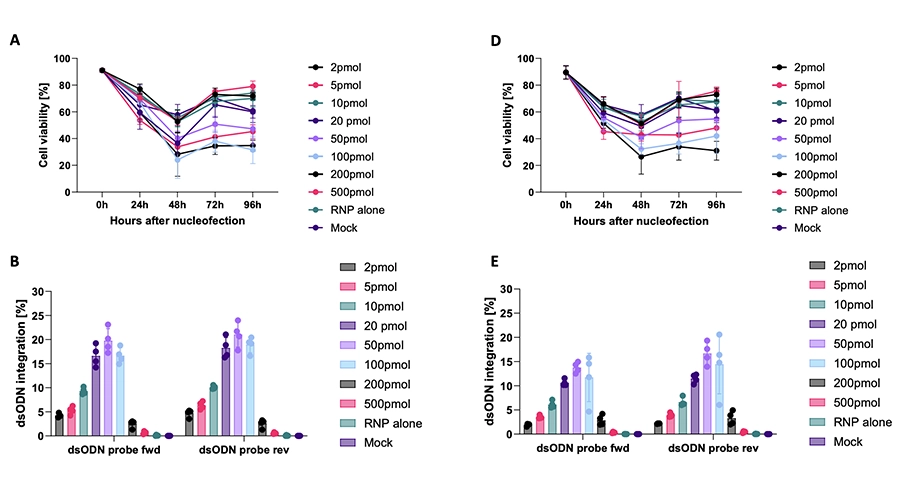
Here, we present a safe and efficient T cell single nucleotide variant (SNV) correction pipeline based on homology-directed repair (HDR), suitable for diverse monogenic mutations. By using founder mutations of Deficiency of ADA2 (DADA2), Autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy (APECED) and Cartilage Hair Hypoplasia (CHH) as IEI models, we show that our pipeline can achieve up to 80% bi-allelic editing, with resultant functional correction of the disease phenotype in patient T cells.
We do not find detectable pre-malignant off-target effects or karyotypic, transcriptomic or proteomic aberrations upon profiling patient T cells with GUIDE-seq, single cell RNA sequencing, PacBio based long-read whole genome sequencing, and high-throughput proteomics.
This study demonstrates that HDR-based SNV editing is a safe and effective option for IEI T cell correction and that it could be developed to an autologous T cell therapy, as the presented protocol is scalable for a GMP-compatible workflow. This study is a step towards the development of gene correction platform that targets a broad number of monogenic mutations.