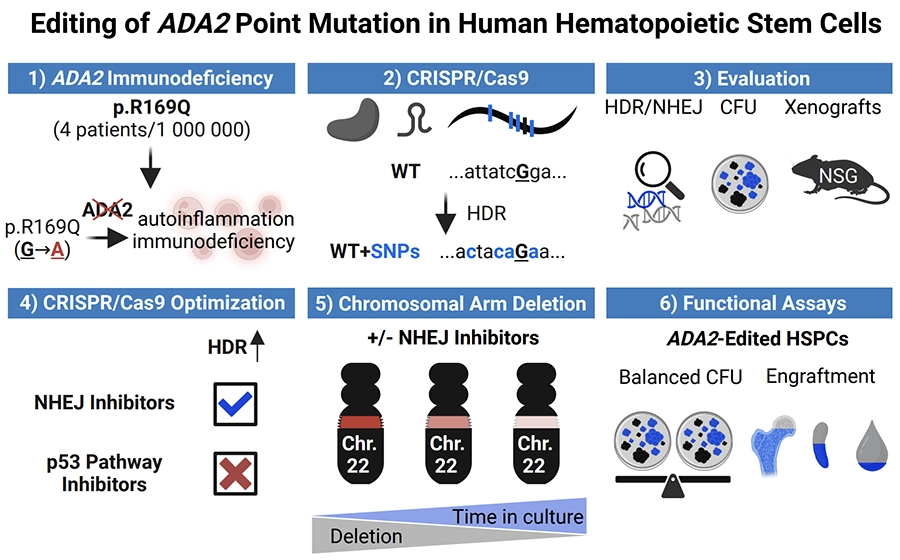
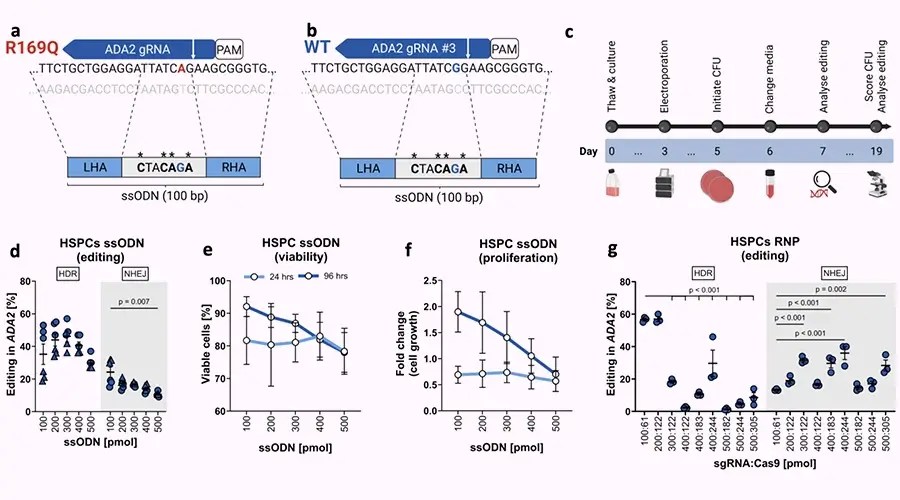
We’re pleased to share our latest preprint, posted on bioRxiv on August 14, 2025: Editing of ADA2 Point Mutation in Human Hematopoietic Stem Cells.
Abstract
Background: The homozygous ADA2: c.506G>A (p.Arg169Gln; p.R169Q) variant accounts for majority of Deficiency in Adenosine Deaminase 2 (DADA2). This monogenic disorder may be amenable to ex vivo gene therapy by correcting the pathogenic mutation in CD34+ hematopoietic stem and progenitor cells (HSPCs).
Objective: To apply CRISPR-Cas9 and homology-directed repair (HDR) as a surrogate strategy to model correction of the pathogenic ADA2 c.506G>A variant in healthy cord blood HSPCs.
Methods: HSPCs were electroporated with optimised CRISPR-Cas9 editing reagents, and editing outcomes, including HDR and on-target deletions, were quantified by ddPCR. Cell functionality was assessed through colony-forming unit (CFU) assays and by xenotransplantation into NOD SCID Gamma (NSG) mice. Two HDR enhancement strategies were tested: (1) genetic inhibitors of p53 and non-homologous end joining (NHEJ) pathways, and (2) pharmacological NHEJ inhibition.
Results: Small-molecule NHEJ inhibitors increased HDR efficiency approximately two-fold (from ∼40 % to ∼80 %). Edited HSPCs retained normal CFU capacity and successfully engrafted in NSG mice. However, up to 8 % of edited cells exhibited on-target chromosome loss, though this declined over time. Up to 40 % of T cells and fibroblasts demonstrated similar losses under NHEJ inhibitors treatment. In contrast, genetically encoded inhibitors did not improve HDR.
Conclusion: The ADA2 p. c.506G>A variant can be effectively edited employing surrogate strategy in HSPCs without impairing functionality. Although pharmacological inhibition of NHEJ enhances HDR efficiency, it also increases the risk of on-target chromosome aberrations, highlighting the need for careful consideration of the associated risks and benefits in therapeutic gene editing.